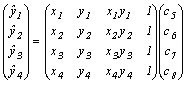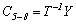This activity is similar to the previous one except that another method is implemented here. Objects are grouped here using probabilistic classification.
Instead of 3, only 2 classes are considered here which are all from Activity 14, Snacku rice crackers and Superthin biscuits. As assembled previously, 10 samples represent each class. These are shown below.
Also, the 4 features which are the sample's pixel area, normalized chromaticity values in red, green, and blue and their corresponding data from Activity 14 are used for probabilistic classification. Each class is again divided into a training set and a test set where each set contains 5 samples.
The probabilistic classification is rooted from the algorithm of linear discriminant analysis (LDA). From the following discriminant function, a test object k is assigned to class i that has maximum f_i.
Note that the transpose matrix operation is represented by T and -1 is the inverse matrix operation. The last term contains the prior probability of class i shown below.
From the equation above, n_i is the number of samples in class i and N is the total number of samples in the training set. Meanwhile, matrix C is called the pooled within class covariance matrix and this obtained from matrix c which is called the covariance matrix. These are as follows.
Note that there are g total number of classes. Covariance matrix is determined from the mean corrected data and the features of the training set which are represented by the following expressions from top to bottom respectively.
The global mean vector mu consists the average of each feature w_i,j,m coming from all classes of the training set and M is the total number of features. Going back to the discriminant function f_i, mu_i is composed of the mean of each feature just from class i in the training set. Thus, using LDA to classify an unknown or test sample, a training set is still required in order to establish the discriminant function which categorize the test sample with the same features possessed by the training set to a class.
The table below summarizes the data gathered from the training set (click the table for larger view).
Finally, the results of the LDA to the test set are summarized in the following table (click the table for larger view).
It can be observed that the values between the two discriminant functions are very small for all samples of the test set compared to the large deviations between the minimum distances acquired in Activity 14. The large values of discriminant function are because of the quantified feature pixel area which is relatively greater than the normalized chromaticity values. Nevertheless, all 10 samples of the test set are correctly grouped to their respective classes.
I grade myself 10/10 in this activity because I have implemented LDA properly and grouped all test samples to their expected classes.
I have successfully finished the activity individually. However, I have helped my classmates on how to handle this activity.
Appendix
The whole Scilab code below is utilized in this activity.
Instead of 3, only 2 classes are considered here which are all from Activity 14, Snacku rice crackers and Superthin biscuits. As assembled previously, 10 samples represent each class. These are shown below.
Also, the 4 features which are the sample's pixel area, normalized chromaticity values in red, green, and blue and their corresponding data from Activity 14 are used for probabilistic classification. Each class is again divided into a training set and a test set where each set contains 5 samples.
The probabilistic classification is rooted from the algorithm of linear discriminant analysis (LDA). From the following discriminant function, a test object k is assigned to class i that has maximum f_i.
Note that the transpose matrix operation is represented by T and -1 is the inverse matrix operation. The last term contains the prior probability of class i shown below.
From the equation above, n_i is the number of samples in class i and N is the total number of samples in the training set. Meanwhile, matrix C is called the pooled within class covariance matrix and this obtained from matrix c which is called the covariance matrix. These are as follows.
Note that there are g total number of classes. Covariance matrix is determined from the mean corrected data and the features of the training set which are represented by the following expressions from top to bottom respectively.
The global mean vector mu consists the average of each feature w_i,j,m coming from all classes of the training set and M is the total number of features. Going back to the discriminant function f_i, mu_i is composed of the mean of each feature just from class i in the training set. Thus, using LDA to classify an unknown or test sample, a training set is still required in order to establish the discriminant function which categorize the test sample with the same features possessed by the training set to a class.
The table below summarizes the data gathered from the training set (click the table for larger view).
Finally, the results of the LDA to the test set are summarized in the following table (click the table for larger view).
It can be observed that the values between the two discriminant functions are very small for all samples of the test set compared to the large deviations between the minimum distances acquired in Activity 14. The large values of discriminant function are because of the quantified feature pixel area which is relatively greater than the normalized chromaticity values. Nevertheless, all 10 samples of the test set are correctly grouped to their respective classes.
I grade myself 10/10 in this activity because I have implemented LDA properly and grouped all test samples to their expected classes.
I have successfully finished the activity individually. However, I have helped my classmates on how to handle this activity.
Appendix
The whole Scilab code below is utilized in this activity.
snacku_training = fscanfMat('snacku_training.txt');
superthin_training = fscanfMat('superthin_training.txt');
snacku_test = fscanfMat('snacku_test.txt');
superthin_test = fscanfMat('superthin_test.txt');
x_training = [snacku_training; superthin_training];
x_test = [snacku_test; superthin_test];
myu1 = [mean(snacku_training(:,1)), mean(snacku_training(:,2)), mean(snacku_training(:,3)), mean(snacku_training(:,4))];
myu2 = [mean(superthin_training(:,1)), mean(superthin_training(:,2)), mean(superthin_training(:,3)), mean(superthin_training(:,4))];
myu = [mean(x_training(:,1)), mean(x_training(:,2)), mean(x_training(:,3)), mean(x_training(:,4))];
x01 = [snacku_training(1,:) - myu; snacku_training(2,:) - myu; snacku_training(3,:) - myu; snacku_training(4,:) - myu; snacku_training(5,:) - myu];
x02 = [superthin_training(1,:) - myu; superthin_training(2,:) - myu; superthin_training(3,:) - myu; superthin_training(4,:) - myu; superthin_training(5,:) - myu];
n1 = 5;
n2 = 5;
c1 = (x01'*x01)/n1;
c2 = (x02'*x02)/n2;
for r = 1:4
for s = 1:4
C(r, s) = (n1/(n1 + n2))*(c1(r, s) + c2(r, s));
end
end
invC = inv(C);
P1 = n1/(n1 + n2);
P2 = n2/(n1 + n2);
P = [P1; P2];
for k = 1:(n1 + n2)
f1(k) = myu1*invC*x_test(k,:)' - (1/2)*myu1*invC*myu1' + log(P(1));
f2(k) = myu2*invC*x_test(k,:)' - (1/2)*myu2*invC*myu2' + log(P(2));
end
f = [f1, f2];
//fprintfMat('f_test.txt', f);
class = [];
for i = 1:(n1 + n2)
[a, b] = find(f(i,:) == max(f(i,:)));
class = [class; b];
end
superthin_training = fscanfMat('superthin_training.txt');
snacku_test = fscanfMat('snacku_test.txt');
superthin_test = fscanfMat('superthin_test.txt');
x_training = [snacku_training; superthin_training];
x_test = [snacku_test; superthin_test];
myu1 = [mean(snacku_training(:,1)), mean(snacku_training(:,2)), mean(snacku_training(:,3)), mean(snacku_training(:,4))];
myu2 = [mean(superthin_training(:,1)), mean(superthin_training(:,2)), mean(superthin_training(:,3)), mean(superthin_training(:,4))];
myu = [mean(x_training(:,1)), mean(x_training(:,2)), mean(x_training(:,3)), mean(x_training(:,4))];
x01 = [snacku_training(1,:) - myu; snacku_training(2,:) - myu; snacku_training(3,:) - myu; snacku_training(4,:) - myu; snacku_training(5,:) - myu];
x02 = [superthin_training(1,:) - myu; superthin_training(2,:) - myu; superthin_training(3,:) - myu; superthin_training(4,:) - myu; superthin_training(5,:) - myu];
n1 = 5;
n2 = 5;
c1 = (x01'*x01)/n1;
c2 = (x02'*x02)/n2;
for r = 1:4
for s = 1:4
C(r, s) = (n1/(n1 + n2))*(c1(r, s) + c2(r, s));
end
end
invC = inv(C);
P1 = n1/(n1 + n2);
P2 = n2/(n1 + n2);
P = [P1; P2];
for k = 1:(n1 + n2)
f1(k) = myu1*invC*x_test(k,:)' - (1/2)*myu1*invC*myu1' + log(P(1));
f2(k) = myu2*invC*x_test(k,:)' - (1/2)*myu2*invC*myu2' + log(P(2));
end
f = [f1, f2];
//fprintfMat('f_test.txt', f);
class = [];
for i = 1:(n1 + n2)
[a, b] = find(f(i,:) == max(f(i,:)));
class = [class; b];
end